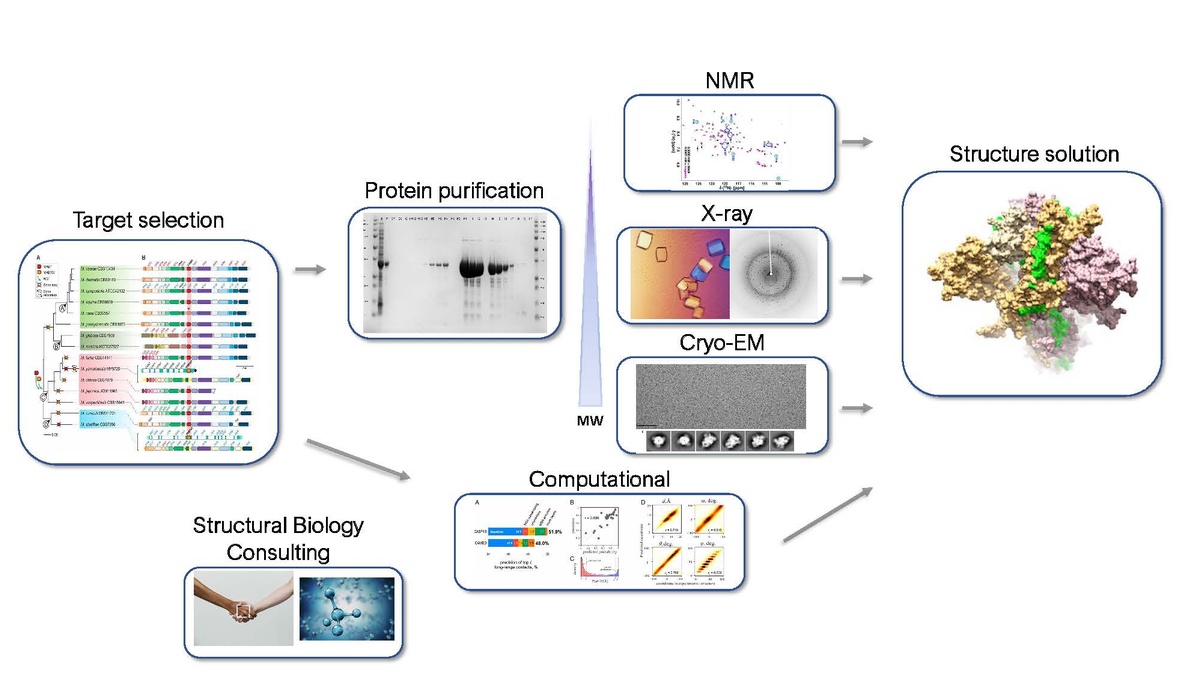
Production Pipeline
The target selection team will use a series of bioinformatic and manual filters to select targets each year for entry into the protein production and structure determination pipeline. Each target will undergo cloning (or synthetic gene design), expression testing, and protein production at UW-PPG or SCRI. Protein from successful targets will be sent to KU for crystallization screening and pre-qualified crystal samples will be sent to the synchrotron beam-lines for X-ray structure solution. A small number of high priority targets from proteins that fail to crystallize will be selected for NMR analysis at WSU and UW-NMR. Large proteins will be solved by Cryo-electron microscopy at UW-CryoEM, SCRI, or KU. In addition, SSGCID structural biology experts offer consultations to the research community.
We anticipate that these approaches will yield 75-100 protein structures per year, including several structures of the same protein(s) with different ligands. Structures will be submitted to the Protein Data Bank (PDB), and materials (clones and protein) generated will be publicly available. Metrics for deposition of X-ray crystal structures into the PDB were previously agreed upon between the two NIAID-funded centers, SSGCID and CSBID (formerly CSGID).