Target Selection
The Target Selection Team will carry out target selection as appropriate each year, usually resulting in batches of 100-300 protein targets. More detailed information about each batch can be found by following the links below. In addition, Target Requests from the scientific community will be reviewed for entry into the Production Pipeline on a monthly basis.
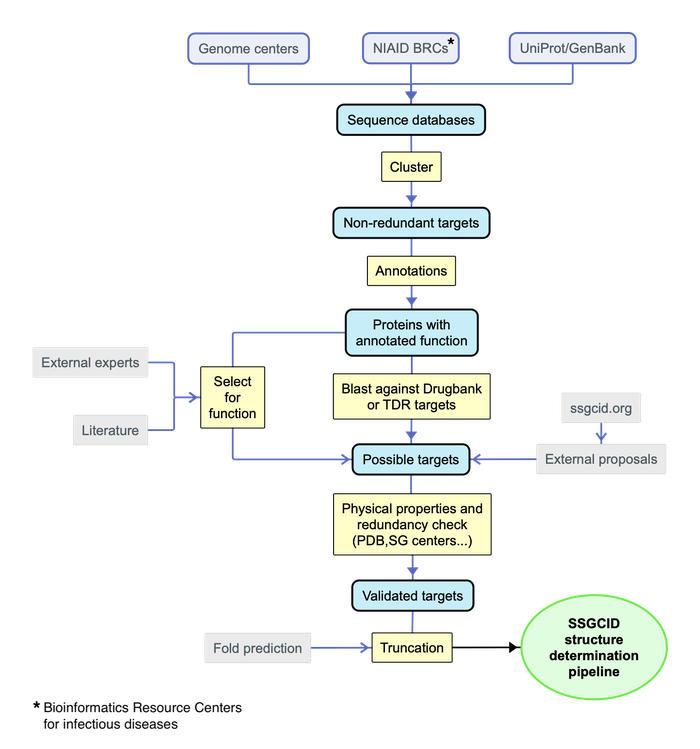
A series of bioinformatic and manual filters will be used to select targets from all the proteins predicted from the genomes of the targeted organisms. These target selection filters will include sequence similarity to known drug targets, documented or potential roles in cell growth, pathogenesis, or drug resistance, as well as markers of infection and vaccine candidates (based on their annotation and/or links to the PubMed database). Those already targeted by other Structural Genomics projects will be excluded. Further prioritization will be assigned on the basis of physical properties (such as predicted solubility, appropriate size, and absence of low complexity sequence). These approaches will be used to select an appropriate number of targets for expression each year (historically ~500 targets/year).