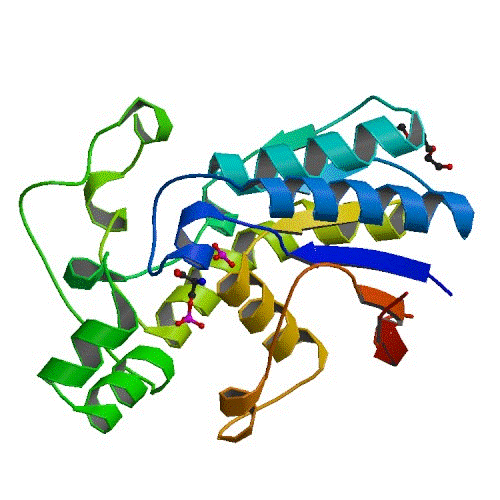
PGAM
- 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase from Burkholderia pseudomallei:
We present here an ensemble of structures solved by the Seattle Structural Genomics Center for Infectious Disease (SSGCID) of 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, or PGAM, from Burkholderia pseudomallei, a pathogen which causes the serious skin infection melioidosis.
The phosphoglycerate mutase family encompasses enzymes that catalyse the interconversion of 3-phosphoglycerate and 2-phosphoglycerate and most organisms utilize PGAM to accomplish this reaction in glycolysis, the metabolic pathway which converts glucose into energy. This protein has been the subject of extensive investigation by X-ray crystallography as three-dimensional crystal structures of PGAM have been solved from bacteria such as Escherichia coli, Thermus thermophiles and Mycobacterium tuberculosis as well as from the fungus Saccharomyces cerevisiae, to name a few.
Here, we present six X-ray crystal structures of PGAM from B. pseudomallei (SSGCID ID BupsA.00114.a) bound to various ligands, including both its substrate (the molecule which PGAM acts upon) and compound fragments which could form the basis for structure-based drug design. The unbound structure (3ezn) consists of a dimer. The structure 3fdz, bound with 3-phosphoglycerate, phosphohistidine and 2,3-bisphosphoglycerate has captured the presence of two distinct stages of the PGAM catalytic cycle. Two vanadate structures were obtained, each with additional molecular entities covalently bound to vanadate (3gw8, vanadate–glycerol complex; 3gp5, vanadate–3-phosphoglycerate complex). Vanadate is a reactive metal oxoanion that is capable of adopting structures that mimic the transition state of phosphoryl-transfer reactions. Together with structure 3fdz, these vanadate structures provide a comprehensive picture of the function of this Burkholderia enzyme. The ensemble of PGAM structures also includes two fragment-bound models (3gp3, depicted in the figure, bound 2-phosphoserine, a compound selected from the Fragments-of-Life™ library (Emerald Bio); 3lnt, bound with malonate). The term ‘fragments’ is used in the context of fragment-based drug discovery, where a fragment is any small molecule of molecular weight approximately less than 300 Da. The structures of PGAM bound to 2-phosphoserine and malonate are examples of non-substrate fragment molecules that may inform inhibitor design.
The ensemble of six X-ray crystal structures of B. pseudomallei PGAM presented here provides for the first time a comprehensive view of a phosphoglycerate mutase enzyme complete with bound ligands encompassing substrate, intermediate, transition-state mimic and fragment-bound structures. Taken together, these crystal structures allow the definitive identification of key amino-acid residues involved in substrate binding and catalysis. The inclusion of two metabolite-like fragment structures in the ensemble (2-phosphoserine and malonate) provides starting points for elucidating ligand-binding flexibility and potential for small-molecule inhibitor design.
These structures were published in the journal Acta Crystallographica Section F. Reference: “An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase. Davies, D., Staker, B., Abendroth, J., Edwards, T., Hartley, R., Leonard, J., Kim, H., Rychel, A.L., Hewitt, S.N., Myler, P.J., and Stewart, L.J. Acta Cryst. (2011) F67, 9:1044-1050 (PDF). PMID: 21904048.