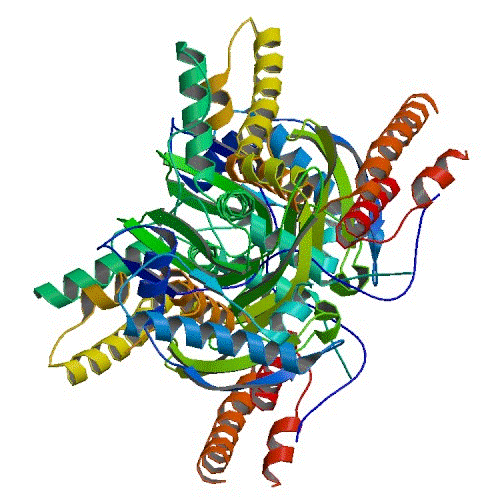
H1N1 PA
The Spanish Flu of 1918 was caused by an H1N1 strain of Influenza A Virus (IAV) and killed 50 to 100 million people worldwide. The threat of a flu pandemic persists with each annual flu cycle and each year hundreds of millions of dollars are spent developing and producing annual flu vaccines. Only limited small molecule drugs are available for treatment of flu infected persons. IAV belongs to a viral class called negative-sense RNA viruses. In order for IAV genes to be translated (made into protein), the negative-sense intermediate RNA strand must be converted to a complimentary positive-sense molecule first. The RNA-dependent RNA polymerase (RdRp) complex of IAV responsible for viral transcription and replication poses an attractive target for drug development.
The Seattle Structural Genomics Center for Infectious Disease (SSGCID) has determined the first structure of the apo c-term domain, a part of the RdRp complex necessary for viral replication, of the polymerase acidic protein, PA. This structure is of PA from the 1933 Wilson-Smith IAV strain (A/WS/1933/H1N1 UniProt ID P15659) spanning the latter 2/3 of the protein (residues 254 through the native C-terminus at 716). The Wilson-Smith 1933 H1N1 strain was the first flu virus cultured in the lab and has been well studied due to its high sequence homology to the 1918 pandemic Spanish flu. The structure reveals a view of residue S552 not previously seen. S552 is a critical residue, mutation of which may allow avian flu strains to bypass the species-species restriction of the avian polymerase complex.
This work is featured in Moen et al., Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1, Sci. Rep. 2014 Aug 4;4:5944 PubMed:25089892. Coordinates are available in the Protein Data Bank www.rcsb.org, PDBID 4IUJ.