SSGCID
Seattle Structural Genomics Center for Infectious Disease
MTB NTM PDB network
Explore the SSGCID Mtb-NTM-PDB network interactively using BioFabric
Supplementary material for Baugh, L et al., Combining functional and structural genomics to sample the essential Burkholderia structome (2014), Tuberculosis, S1472-9792(14)20565-8. PMID: 25613812.
How to load the network into BioFabric:
- Download the SSGCID biofabric network data file mtb_ntm_pdb_network.bif
- Launch BioFabric (requires Java), or download and install your own copy from biofabric.org
- Use the top menu File > Load from File to load the network data file
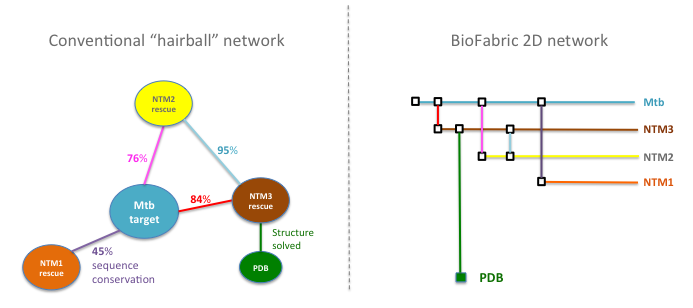
Schematic comparisons betwen conventional network (left) and 2D network view (right), where targets (nodes) are represented as horizontal lines (with Mtb targets arrayed across the top and NTM targets below) connected by vertical lines representing BlastP matches with >40% amino acid similarity. Targets with structures submitted to the PDB appear as long vertical line.