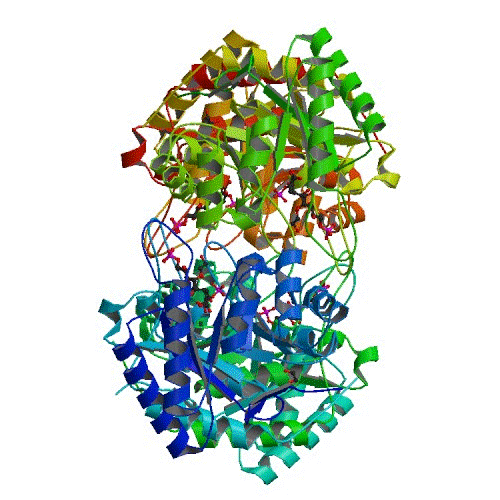KDOP
Gram-negative bacteria are emerging as a threat to humans, in that they have become resistant to all the available antibiotics used to treat humans. Thus many organizations, such as the World Health Organization and the Infectious Diseases Society of America, are highlighting this crisis and are calling for increased effort to find novel antibiotics directed against Gram-negative bacteria.
In one part of its effort to elucidate new anti-bacterial drug targets, Seattle Structural Genomics Center for Infectious Disease (SSGCID) has solved three new structures for a promising antibacterial drug target, bacterial KDOP synthases (aka 3-deoxy-D-manno-2-octulosonate-8-phosphate synthase), from a diverse group Burkholderia bacteria. Burkholderia are a group of gram-negative bacteria that cause severe, life threatening infections, including melioidosis, a systemic illness common in SE Asia and of concern for Biodefense of the USA, because of its extreme infectiousness, rapid fatality, and the concern that terrorists might easily grow it and infect people in the USA. Antibiotics to treat Burkholderia are very limited, because of its inherent resistance to commonly used antibiotics, and many lives are lost due to melioidosis every year for this reason. Burkholderia bacteria also cause severe lung infections in children with cystic fibrosis. In children with cystic fibrosis, infections with Burkholderia bacteria frequently become the cause of death because their Burkholderia are resistant to all available antibiotics. Having a new antibiotic against Burkholderia and related bacteria would be life saving for these children.
KDOP synthases are hypothesized to be a great drug target for Gram-negative bacteria and thus also for Burkholderia. These KDOP synthases are an essential step in bacterial outer membrane synthesis, thus are required for bacterial life. They work in the biochemical pathway to create a molecule called lipopolysaccharide (LPS), and LPS is an essential component in the outer membrane of Gram-negative bacteria. Thus an inhibitor of KDOP synthase would be expected to kill Burkholderia and other Gram-negative bacteria. The structures we have solved will facilitate “structure-based drug development” (SBDD). SBDD is a process where a 3-D structure is used to find and improve drug-like inhibitors to become a drug, in this case an antibiotic that is effective against Burkholderia and other Gram-negative bacteria.
“Selective” antibiotics are important in making antibiotics that do not harm humans, but still kill bacteria. There is no LPS in humans. Thus, KDOP synthases are not similar to any human enzymes, and this information can be used to create selective antibiotics, in that inhibiting KDOP synthases will kill bacteria, but will not inhibit human enzymes.
We solved KDOP synthetase structures for three different Burkholderia bacterial species: from Burkholderia cenocepacia (the Burkholderia species that most commonly infects children with cystic fibrosis, PDB.org: 3TML), and Burkholderia pseudomallei (the Burkholderia species that causes melioidosis, PDB.org: 3SZ8, 3UND, and 3TMQ), and Burkholderia ambifaria (the second-most common species of Burkholderia that infects children with cystic fibrosis, PDB: 3T4C), The KDOP synthetase structures have a high degree of structural similarity. Thus we expect that one antibiotic drug, inhibiting all the Burkholderia KDOP synthases, can be made to inhibit all of the Burkholderia bacteria. In fact, since KDOP synthases are similar in all Gram-negative bacteria, it is very likely that an inhibitor will be an effective antibiotic for all Gram-negative bacteria. We have recently proposed that these proteins be the focus of further functional studies to test whether known KDO synthase inhibitors restrict the activity of these proteins and reduce viability of Burkholderia cells.
These structures were published in the journal PLoS ONE. Reference: Combining Functional and Structural Genomics to Sample the Druggable Bacterial Structome. Baugh, L., Gallagher, L.A., Patrapuvich, R., Clifton, M., Gardberg, A.S., Edwards, T.E., Armour, B., Begley, D.W., Dieterich, S.H., Dranow, D.M., Abendroth, J., Fairman, J.W., Foxx, D.III, Staker, B.L., Phan, I., Gillespie, A., Choi, R., Nakazawa-Hewitt, S., Nguyen, M.T., Napuli, A., Barrett, L., Buchko, G.W., Stacy, R., Myler, P.J., Stewart, L.J., Manoil, C., and Van Voorhis, W.C. PLoS One (2012) 8(1):e53851. PMID: 23382856. This article has been recommended by Faculty of 1000.