Prokaryotic type IV secretion systems
Typically comprised of ~12 different scaffold components, prokaryotic type IV secretion systems (T4SSs) translocate a variety of substrates across the cell envelope. Due to the ability to translocate large DNA segments (i.e., mobile genetic elements), some T4SSs contribute to the spread of antimicrobial resistance and virulence genes. Other T4SSs translocate smaller DNA segments and/or proteins into eukaryotic cells (e.g., arthropod, plant and human) in order to benefit bacterial survival. At least eight major groups of T4SSs are described and each group consists of minor variations on a common structural theme. Within a given bacterial genome, combinations of different T4SS groups can be encoded, and sometimes multiple copies of the same T4SS group can be present. As an example, two functionally divergent T4SSs (vir and trw) are found in many species of Bartonella, bacteria that are transmitted by ticks and fleas and cause more than 20,000 of a range of diseases in Americans annually, including endocarditis. In species of Rickettsia, which are also spread by ticks and fleas and cause various fevers and typhus, only one T4SS exists, although many of the scaffold components are encoded by multiple divergent gene copies. Curiously, little is known regarding the ability for cross-system interchangeability of scaffold proteins between different T4SSs encoded within the same bacterial genome. What is particularly puzzling is how distinct T4SSs achieve correct scaffold assembly without interfering with one another throughout the bacterial lifecycle.
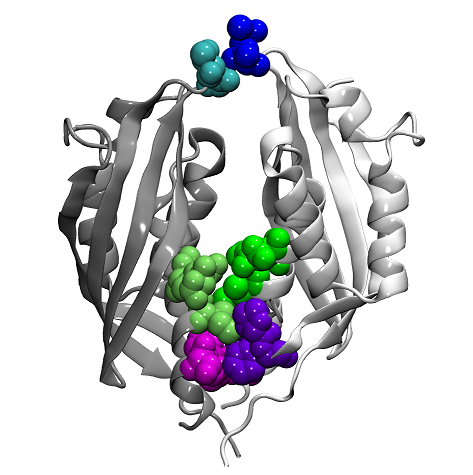
In a collaboration between Drs. Joseph Gillespie (University of Maryland Baltimore), Arto Pulliainen (University of Helsinki, Finland and University of Turku, Finland), Holger Scheib (University of Queensland, Australia) and the Seattle Structural Genomics Center for Infectious Disease (SSGCID), a comparative structural analysis was undertaken using crystal structures for six proteins solved by the SSGCID and a modeled structure for a seventh protein. The proteins studied are from the T4SS VirB8 family of channel proteins from several pathogenic species of Bartonella and Rickettsia typhi, the agent of murine typhus. The Bartonella proteins belong to two functionally different T4SS classes (the highly divergent vir and trw). The R. typhi proteins, RvhB8-I and RvhB8-II, are divergent duplications that are assumed to assemble into one T4SS, and this study presents the first crystal structure solved for one of these rvh proteins.
Together with results from biochemical assays, protein modeling and bioinformatics, the researchers investigated the factors regulating maintenance of multiple T4SSs components within single bacterial species. The data suggest that two distinct mechanisms exist that prevent cross-system interchangeability between different T4SSs (Bartonella species) or divergent scaffold duplicates (Rickettsia species) within the same bacterium. For Bartonella species, spatiotemporal regulation was shown to form a barrier, while in Rickettsia species, structural barriers due to rapid diversification were found. This work has potential to impact therapeutics developed on T4SSs, which are ideal drug targets provided that no analogous counterparts are known from eukaryotes. Drugs targeting the barriers to cross-system interchangeability (i.e. regulators) could dysregulate the structural and functional independence of discrete systems, potentially creating interference that prevents their efficient coordination throughout bacterial infection.
Details of this work have been published in the journal mBio. Reference: Structural insight into how bacteria prevent interference between multiple divergent type IV secretion systems. Gillespie, J.J., Phan, I.Q.H., Scheib, H., Subramanian, S., Edwards, T.E., Lehman, S. S., Piitulainen, H., Rahman, M.S., Rennoll-Bankert, K.E., Staker, B.L.,Taira, S., Stacy, R., Myler, P.J., Azad, A.F., Pulliainen, A.T. mBio 6(6):e01867-15 (PubMed ID 26646013). Coordinates are available in the Protein Data Bank www.rcsb.org, PDBIDs 4O3V;4NHF; 4LSO; 4KZ1; 4MEI; 4JF8